Abstract
T-cell large granular lymphocyte leukemia (T-LGLL) is a rare disease characterized by clonal proliferation of CD3+ lymphocytes whose pathogenesis is not completely understood. LGL resistance to the activation-induced cell death is sustained by anti-apoptotic pathways, mainly due to constitutive activation of the JAK-STAT axis by activating somatic mutations of STAT3 or STAT5B genes or epigenetic disturbances. STAT3 mutations are peculiar to CD8+ T-LGLL and are associated with cytopenias, treatment requirement, and reduced survival. STAT5B lesions are mostly found in CD4+ T-LGLL, a disorder presenting with a mild clinical course. A similar behaviour has been observed also in both STAT wild type (WT) CD8+ and CD4+ forms. The link between STAT3-dependent miR-146b repression and neutropenia exemplifies the role that the pathogenetic effect of somatic mutations can exert through complex axes, also involving non-coding RNAs. Since in the most symptomatic form of LGLL an urgent need to discover new mechanisms in place is demanded, we focused on additional regulatory molecules such as circular RNAs (circRNAs), single-stranded covalently closed RNA molecules generated by backsplicing (Figure 1A). CircRNAs' oncogenic potential is linked to their activity as miRNA sponges, modulators of RNA-binding protein activity, and their ability to encode oncogenic peptides.
The study group includes 20 patients recruited at the Hematology Unit of Padua University Hospital (Italy) and diagnosed with T-LGLL according to the WHO criteria, plus 5 healthy controls (CTR). Patient leukemic clones were equally distributed among CD8+ WT and STAT3-mutated and CD4+ WT and STAT5B-mutated. RNA-seq of ribodepleted RNA from immunomagnetically purified CD57+ leukemic clones and CD8+ cells from CTR was obtained to study circRNAs. CircRNAs were identified and quantified from RNA-seq data by CirComPara v0.6.3. Differential expression was assessed by edgeR with robust estimation of dispersion. The circular to linear proportion (CLP), indicating the relative abundance of each circRNA with respect to all the overlapping host gene transcripts, was calculated, and differential CLP across conditions was assessed with CircTest.
We detected 68.619 circRNAs and focused on 5.948 with high expression. These were expressed from 2,940 loci, mostly from annotated genes (99.95%). We identified 28 circRNAs derived from genomic regions without known genes. About one-half of circRNA host genes expressed multiple, up to 20, circular isoforms. About 4% of circRNAs had a CLP of at least 0.25, with 95 circRNAs more expressed than the linear counterpart, including circGUSBP2, circSHOC1, circTCEANC, circZNF609, and circKLHL8.
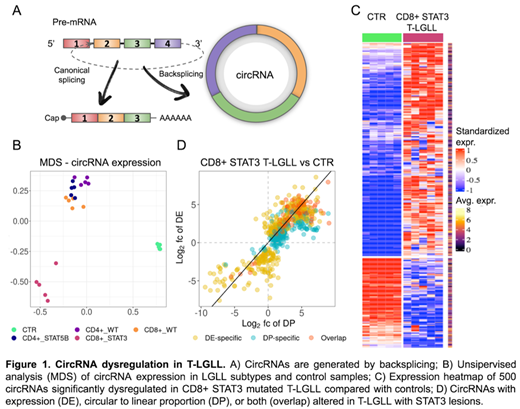
Unsupervised analysis of circRNA expression showed a clear separation of T-LGLL from CTR samples, pointing toward circRNA dysregulation in T-LGLL, particularly for the CD8+ STAT3 mutated group, which diverged from the other subtypes (Figure 1B). The direct comparison between the CD8+ STAT3-mutated, the three molecular subtypes as a single group (OTH), and the CTR samples identified differences in circRNA expression specifically linked to the most symptomatic group with STAT3 lesions and those commonly observed in T-LGLL. In STAT3-mutated, 500 circRNAs were dysregulated with an excess (71%) of upregulation compared to CTR (Figure 1C). Most circRNAs commonly altered in both LGLL groups had a more marked dysregulation in patients with STAT3 mutations, as observed for circIKZF2 and a newly discovered intergenic circRNA. Most circRNAs with a significantly varied CLP (96%) showed an increased expression of the circular than linear transcripts in T-LGLL compared to CTR, uncovering genes with an imbalance of circular to linear transcripts proportion in leukemic cells (Figure 1D).
The ongoing validation of the differential circRNA expression in an extended cohort of 40 LGLL patients is prioritizing circRNAs for functional investigation. The integration of computational predictions and in vitro functional screening through specific circRNA silencing will attempt to understand the link between circRNAs dysregulated in T-LGLL and disease mechanisms, unveiling possible circRNA-involving axes governed by hyperactivating STAT3 somatic variants that contribute to the malignant LGL phenotypes.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal